Original Brief
Tem and CTX-M extended-spectrum beta-lactamase in Klebsiella spp and Escherichia coli isolates from inanimate surfaces of hospital environments
Marco Rivera-Jacinto1,a, Claudia Rodriguez-Ulloa1,b, Rene Flores Clavo2,c, Luis Serquen Lopez 2,d, Zhandra Arce Gil 3,e
1 Faculty of Health Sciences, Universidad Nacional de Cajamarca. Cajamarca, Peru
2 Regional Hospital of Lambayeque. Lambayeque, Perú.
3 Universidad Católica Santo Toribio de Mogrovejo. Lambayeque, Peru.
a Biologist-microbiologist, doctorate in biomedical sciences; b biologist-microbiologist, doctorate un biomedical sciences; c licentiate in microbiology; dlicentiate in microbiology; elicentiate in microbiology, master’s degree in sciences.
ABSTRACT
The aim of the study was to determine the genotype of 15 ESBL strains of Enterobacteriaceae resistant to beta-lactams, isolated from inanimate surfaces and phenotypically characterized as producing extended-spectrum beta-lactamase. After evaluation and screening of the bacterial strains, a PCR was conducted to amplify fragments of 1078 bp and 544 bp corresponding to type TEM and CTX-M ESBL. Eleven strains presented both fragments at the time and only three had blaCTX-M. In conclusion, the presence of ESBL genes in cultures from the environment was demonstrated, some of which may belong to more than one type. This information could serve as a basis for implementing preventive measures to prevent the transmission of multiresistant bacteria from inanimate surfaces to patients, mainly in critical hospital areas.
Key words: beta-Lactamases; Cross infection; Klebsiella pneumoniae; Escherichia coli; Polymerase chain reaction (source: MeSH NLM).
INTRODUCTION
β-Lactamases are bacterial enzymes that can hydrolyze antibiotics β-lactams, thus making them harmless for bacteria. Extended spectrum β-lactamases (ESBL) are among those of greater clinical relevance and include three main types: TEM, SHV, and CTX-M (1). TEMs come from the classic TEM-1 and TEM-2 that are present in plasmids, whereas SHVs have a chromosomal origin (1,2). CTX-Ms, which have a different evolutionary history, have intrinsic ESBL activity derived from its chromosomal predecessor in Kluyvera (2) and are the most widely reported ESBLs worldwide, predominantly in pathogens of nosocomial infections (NI) such as Escherichia coli and Klebsiella spp. (2-4).
Although the endogenous source is the most important one, the hospital environment is a possible reservoir of these pathogens (5,6) because inanimate surfaces can be colonized by ESBL-producing bacteria (7,8) and serve as a potential source of cross-contamination and colonization of the hands of healthcare workers. The objective of this work was to determine the ESBL genotype of 15 strains of Enterobacteriaceae that were isolated from inanimate surfaces, phenotypically characterized as ESBL producers in a preliminary study at the Regional Hospital of Cajamarca (7) in north Peru.
THE STUDY
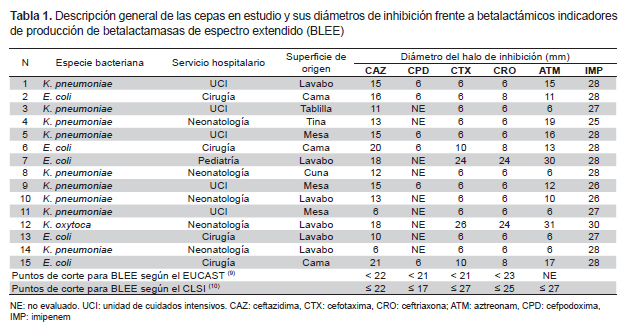
The 125 samples were collected by swabbing sinks, tables, beds, and other surfaces in operating rooms, the pediatric ward, medicine ward, neonatology ward, maternity ward, and intensive care units (ICUs), from September 2009 to June 2010. Of 20 strains of E. coli, 19 of K. pneumoniae, and 4 of K. oxytoca, 15 strains (5 of E. coli, 9 of K. pneumoniae, and 1 of K. oxytoca) were previously found to be ESBL producers by phenotypic tests (double disc and combined disc synergy) in a preliminary study (7) and were reactivated and subjected, once more, to screening tests with β-lactam indicators using the cut-off points proposed by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (9) and the Clinical and Laboratory Standards Institute (CLSI) (10).
DNA extraction from the bacteria of each strain was performed with the Pure Link Genomic DNA Kit (Invitrogen), as per the protocol for bacterial samples. The PCR primers were as follows: blaTEM forward 5′-ATAAAATTCTTGAAGACGAAA-3′, reverse 5′-GACAGTTACCAATGCTTAATC-3′ and blaCTX-M forward 5′-TTTGCGATGTGCAGTACCAGTAA-3′ and reverse 5′-CGATATCGTTGGTGGTGCCAT-3′, and were used to amplify fragments of 1078 bp of TEM (11) and of 544 bp of CTX-M (12). The reaction mixture was adjusted to the final volume of 50 µL for amplification by means of Fast Start Taq DNA Polymerase (Roche): 25.6 µL PCR-grade water; 5 µL of 10× buffer, 4 µL 25 mM MgCl2, 1 µL of the nucleotide mix; 5 µL of each primer; 0.4 µL of Taq DNA polymerase, and 4 µL of extracted DNA. The PCR was performed on a Verita Thermal Cycler (Applied Biosystems) under the previously described conditions (11,12).
Electrophoresis was performed at 100 V for 45–60 minutes in a 1.5% agarose gel (Promega, USA) with ethidium bromide (1.0 mg/mL); the products were visualized by means of a Bio-Rad Pharos FX Plus® transilluminator, and the images were analyzed in the Quantity One software (Bio-Rad, USA). The size of the amplified DNA was verified using a molecular-weight marker of 1500 bp.
FINDINGS
All the strains tested with ceftazidime (30 μg) and cefpodoxime (10 μg) showed smaller inhibition diameters than those indicated by the EUCAST and CLSI as cutoff points for ESBL screening. Strains 7 (E. coli) and 12 (K. oxytoca) showed greater cutoff diameters for cefotaxime (30 μg) and ceftriaxone (30 μg) according to the EUCAST but smaller than those indicated by the CLSI; these same strains yielded halos greater than 30 mm for aztreonam (30 μg). During imipenem testing (10 μg), all strains showed halos greater than the cutoff according to the EUCAST for carbapenemase (Table 1).
Seven strains of K. pneumoniae and 4 strains of E. coli carried both genes (blaTEM and blaCTX-M), 2 strains of K. pneumoniae and 1 strain of K. oxytoca had only blaCTX-M, and a strain of E. coli isolated from the pediatric ward did not have the genes we used for testing (Figures 1 and 2).
DISCUSSION
Various studies showed that K. pneumoniae is found at a greater frequency than E. coli in the hospital environment, including inanimate surfaces (5,6,8). These data are in agreement with the present report (Table 1) because of Klebsiella spp.’s greater ability to survive on surfaces; this trait is important because it may lead to colonization of patients, at a greater or lower frequency, depending on the species of bacteria and their essential biological differences (5,6,8). The high contagiousness of the ESBL-producing bacteria has also been demonstrated in these studies and explains the virtuous circle and vicious circle whereby inanimate surfaces are contaminated by infected patients or by colonized healthcare personnel, who in turn become contaminated via these same surfaces (6). This problem is exacerbated when these microorganisms express genes of resistance to multiple antimicrobials, turning the hospital environment into a reservoir for multidrug-resistant bacteria difficult to eradicate, despite the most thorough cleaning procedures used by hospitals (6).
Of the 15 strains studied, 14 had the ESBL phenotype CTX-M and 11 had the ESBL genotypes CTX-M and TEM (Figures 1 and 2). The greater frequency of the first coincides with a greater frequency of reporting worldwide (2,3). This coincidence is important due to the wide diversity and fast dissemination corresponding to this ESBL during the NI. Some authors stated that it is common among CTX-M-producing bacteria to have different resistance phenotypes (2). In the present study, 13 of 14 strains showed pronounced activity of cefotaximase in the screening test and only one K. oxytoca strain yielded a different pattern; in addition, the high activity of ceftazidimase of the gene CTX-M is noteworthy in strains 11, 12, and 14 (Table 1), which were negative for the TEM gene. The resistance phenotype where cultures show both ESBL types is an increasingly common and dangerous bacterial coproduction strategy among Klebsiella and E. coli strains; this approach sometimes allows these bacteria to inactivate other groups of antimicrobials at high frequency and effectiveness (4). In the case of culture 7, which did not have any of the genes used for testing but showed an ESBL-producing phenotype during the screening, its genotype may belong to the VHS group or an inducible chromosomal gene called AmpC, which is difficult to detect in some cases. These mechanisms may also be responsible for the resistant phenotype of the bacteria with genotype TEM and CTX-M; however, to ensure that we are dealing with a specific type of ESBL TEM or ESBL CTX-M, it is necessary to sequence the genes (not done here). This is an important limitation of this study.
In addition to the presence of the
ESBL-positive bacterial reservoir, the difficulties with its control and treatment are also important problems because of the
ease with which these and other resistance genes are disseminated among
Enterobacteriaceae
(1,4).
Although in the present study, no tests were performed to determine the pattern
of clonality of the samples, it is important to mention that genetic relations
among the selected strains could have been identified by such methods as pulsed
field electrophoresis. This approach should help to determine how many clones
circulate in hospitals or point to possible transmission foci. We would like to
draw the attention of healthcare authorities to our results, in order to
emphasize the monitoring of transmission of these bacteria and other nosocomial
pathogens from the environment, especially from potentially contaminated
inanimate surfaces in critical hospital areas. In this regard, it would also be
useful to establish programs for detection of ESBL and other resistance
mechanisms both at the hospital level and in other scenarios, which is seldom
done in Peru. Author contributions: MRJ and CRU participated in the conception of the study, collection of
data, analysis and interpretation of data, writing, critical review, and
approval of the final
version of the manuscript. RFC, LSL, and ZAG participated in the collection of
data, analysis and interpretation of data, critical review of the manuscript, in
addition to approval of the final version of the manuscript. Funding sources:
The PCR was performed with materials and equipment
from the Molecular Biology Laboratory of the Research Division of the Regional
Hospital of Lambayeque. Conflicts of interest: The authors declare that they do not have conflicts of interest. References 1. Rivera A, Larrosa N, Mirelis B, Navarro F. Importancia de
los controles de calidad para la detección de la resistencia a antibióticos β-lactámicos
en enterobacterias. Enferm Infecc Microbiol Clin 2014;32(Supl 1):30-6. doi:
10.1016/S0213-005X(14)70147-8. 2. D’Andrea MM, Arena F, Pallecchi L, Rossolini GM. CTX-M-type
β -lactamases: A successful story of antibiotic resistance. Int J Med Microbiol
2013;303(6-7):305-17. doi: 10.1016/j.ijmm.2013.02.008. 3. Guzmán-Blanco M, Labarca JA, Villegas MV, Gotuzzo E, On
behalf of the Latin America Working Group on Bacterial Resistance. Extended
spectrum -lactamase producers among nosocomial Enterobacteriaceae in Latin
America. Braz J Infect Dis 2014; 18(4):421–33. doi: 10.1016/j. bjid.2013.10.005.
4. Kassakian SZ, Mermel LA. Changing epidemiology of
infections due to extended spectrum beta-lactamase producing bacteria.
Antimicrob Resist Infect Control. 2014;3(1):9. doi: 10.1186/2047-2994-3-9.
5. Freeman JT, Nimmo J, Gregory E, Tiong A, De Almeida M,
McAuliffe GN, et al. Predictors of hospital surface contamination with Extended-spectrum
β-lactamase-producing Escherichia coli and Klebsiella pneumoniae: patient and
organism factors. Antimicrob Resist Infect Control 2014;3(1):5. doi:
10.1186/2047-2994-3-5. 6. López-Cerero L. Papel del ambiente hospitalario y los
equipamientos en la transmisión de las infecciones nosocomiales. Enferm Infecc
Microbiol Clin. 2014;32(7):459–64. doi: 10.1016/j.eimc.2013.10.004. 7. Rivera-Jacinto MA. Betalactamasas de espectro extendido en
cepas de Escherichia coli y Klebsiella sp aisladas de reservorios inanimados en
un hospital del norte del Perú. Rev Esp Quimioter 2012;25(2):161-63. 8. Guet-Revillet H, Le Monnier A, Breton N, Descamps P,
Lecuyer H, Alaabouche I, et al. Environmental contamination with extended-spectrum
b-lactamases: Is there any difference between Escherichia coli and Klebsiella
spp? Am J Infect Control 2012;40(9):845-8. doi: 10.1016/j.ajic.2011.10.007.
9. European Society of Clinical Microbiology and Infectious
Diseases. EUCAST guidelines for detection of resistance mechanisms and specific
resistances of clinical and/or epidemiological importance. Version 1.0, 2013.
Disponible en:
http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_v1.0_20131211.pdf
10. Clinical and Laboratory Standards Institute (CLSI).
Performance Standards for Antimicrobial Susceptibility Testing; 22th
Informational Supplement. CLSI Document M100-S22. Wayne, PA: CLSI, 2012. 11. Mabilat C, Goussard S. PCR detection and identification
of genes for extendedspectrum β-lactamases. In: DH Persing, Smith TF, Tenover FC,
White TJ, editors. Diagnostic molecular microbiology: principles and
applications. Washington, D.C: American Society for Microbiology; 1993: p.
553-559. 12. Arce-Gil Z, Llontop-Nuñez J, FloresClavo R,
Fernández-Valverde D. Detección del gen CTX-M en cepas de Escherichia coli
productoras de β-lactamasas de espectro extendido procedentes del Hospital
Regional de Lambayeque; Chiclayo-Perú: Noviembre 2012-Julio 2013. Rev. cuerpo
med HNAAA 2013;6(4):13-16. Correspondence: Marco Antonio Rivera Jacinto Address: Av. Atahualpa 1050 – Edificio 1D. Lab.1D-105. Ciudad
Universitaria,Universidad Nacional de Cajamarca,
Cajamarca-Perú. Phone number: 511-976996558 E-mail : mrivera@unc.edu.pe; marco_riverajacinto@yahoo.es Received: 5/7/2015 Approved: 9/2/2015