María V. Méndez
Ysvette Vásquez
Dayana Requena
10.17843/rpmesp.2020.372.4652
ORIGINAL ARTICLE
Typification of the staphylococcal chromosome cassette of methicillin-resistant Staphylococcus aureus in the state of Aragua, Venezuela
Betsi Bastidas
![]() 1, Master of Veterinary Medicine
1, Master of Veterinary Medicine
María V. Méndez
![]() 1, Doctor of Science
1, Doctor of Science
Ysvette Vásquez
![]() 2, Master of Veterinary Medicine
2, Master of Veterinary Medicine
Dayana Requena ![]() 3, Master of Biomedical Sciences
3, Master of Biomedical Sciences
1 Escuela de
Bioanálisis, Facultad de Ciencias de la Salud, Universidad de Carabobo,
Aragua, Venezuela.
2 Laboratorio de Bacteriología, Hospital de los Samanes, Aragua,
Venezuela.
3 Instituto de Investigaciones Biomédicas Dr. Francisco Triana,
Universidad de Carabobo, Aragua, Venezuela.
ABSTRACT
Objective: Typify the SCCmec cassette in methicillin-resistant strains of Staphylococcus aureus in clinical isolates from health centers in the State of Aragua-Venezuela and compare the presence of SCCmec genotypes among the state health centers and according to the type of infection.
Materials and methods: 81 MRSA strains from four health centers of the Aragua-Venezuela State were studied. Methicillin resistance was performed with the Kirby-Bauer method with oxacillin (1 µg) and cefoxitin (30 µg) disks. The mecA gene and SCCmec were analyzed by the multiple PCR technique.
Results: Only 55 isolates (67.9%) amplified the mecA gene, and 24 strains (43.6%) amplified SCCmec. SCCmec type I was the most frequency, followed by SCCmec IV and SCCmec III, representing 62.5%, 25% and 12.5%, respectively. SCCmec I was predominant in health center A (80%), while in B and C 60% and 100% respectively were SCCmec IV. At health center D, 50% turned out to be SCCmec I and 50% SCCmec IVd. A relationship was found between the SCCmec and the health center with statistical significance. SCCmec I predominated in skin and soft tissue and respiratory infections with 63.2% and 50%, respectively. There was no association between genotype and type of infection with a p value greater than 0.05.
Conclusions: The prevalence of SCCmec I and IV will allow establishing new measures in the use of antibiotics and epidemiological control.
Keywords: Staphylococcus aureus; SCCmec; mecA gene; Molecular Epidemiology; Staphylococcus aureus methicilin resistent (source: MeSH NLM).
INTRODUCTION
Methicillin-resistant Staphylococcus aureus
(MRSA) is a global public health problem, causing serious infections in
hospitals and the community. In 2018, the World Health Organization (WHO)
estimated that patients with MRSA infections are 64% more likely to die than
patients with non-resistant infections (1). In addition, by 2017 the
WHO included MRSA in the list of the twelve most dangerous pathogens to human
health because of its resistance to antibiotics (2).
Resistance to methicillin is caused by the fact
that the bacteria synthesizes a penicillin-binding protein known as PBP2a,
which has a low affinity for methicillin and the rest of the beta-lactam
antibiotics, preventing the entry of this type of antibiotic into the bacterial
cell to exert its antimicrobial effect. PBP2a is encoded by the mecA gene, which is found within a mobile
chromosomal element, called the staphylococcal chromosome cassette (SCCmec). The mecA
gene is distributed in both S. aureus and other methicillin-resistant
coagulase-negative staphylococcus species (3,4).
The SCCmec can
measure between 21 and 67 Kb, and has a set of genes such as the ccr (ccrAB and ccrC) that encode recombinases, in addition to the mec complex that contains the mecA
gene, its regulatory genes (mecI, mecR), the acquired genetic determinants, which are
produced as a result of the integration of plasmids and transposons, and
finally, the sequence of the J region (5). It is important to know
the constitution of the SCCmec because according to
recombination events between the ccr and mecA genes, a variety of SCCmec
are generated, and these, allow the classification of MRSA according to the SCCmec it possesses. Initially, five types of SCCmec (I-V) and a number of variants or subtypes (5,6) were described; however, new types were recently
published as SCCmec VI-XI (7).
Furthermore, types of SCCmec
differ from each other because of their resistance determinants. Therefore, SCCmec I, IV, V, VI and VII code for resistance to
beta-lactam antibiotics only, whereas SCCmec
II, III and VIII have additional genes for resistance to multiple classes of
antibiotics other than beta-lactam antibiotics (5,8).
On the other hand, MRSA strains can be
contracted from the hospital environment (MRSAH) or in the community (MRSEC).
MRSAC is characterized by sensitivity to multiple antibiotics and is usually
resistant to beta-lactam antibiotics only. It can cause skin and soft tissue
infections, including severe cases of necrotizing pneumonia, necrotizing
fasciitis, septic thrombophlebitis, and sepsis (9,10). MRSAH is
resistant to several groups of antibiotics, in addition to beta-lactams, and is
associated with patients with risk factors such as high antibiotic consumption,
prolonged hospital stays, invasive procedures (intravenous catheters, urinary
catheters, tracheotomy), bedsores, severe illness, and contact with MRSA
colonized patients (9,10). In addition, MRSAC carries SCCmec IV and V (5), while MRSAH strains
have SCCmec I, II or III (11).
The molecular typification
of SCCmec is performed by polymerase chain
reaction (PCR), using the multiple PCR technique, which allows different types
of SCCmec to be determined simultaneously,
which is very useful in epidemiological studies (12,13). Acuña et al (14) applied the multiple PCR
technique to typify SCCmec in 21 MRSA strains
isolated in the bacteriological laboratory of a hospital in Cumaná,
state of Sucre, where they found SCCmec I and
IV in outpatients and in adult emergency patients. The presence of SCCmec IV genotypes indicated that the bacteria
isolated came from the community and were spreading to hospital services,
producing nosocomial infections. In the state of Zulia, González et al (15)
characterized the SCCmec of 54 MRSA strains by
multiple PCR and showed that 54% had SCCmec
IV; 40%, SCCmec I; while 4% and 2%, SCCmec IA and SCCmec
IIIB, respectively.
In the state of Aragua, no previous studies
have determined the type of SCCmec circulating
in the health centers of the region. This is the reason why the objective of
the present investigation was to typify SCCmec
in MRSA strains isolated from health centers in Aragua.
|
KEY MESSAGES |
|
Motivation for the study:
Methicillin-resistant Staphylococcus aureus (MRSA) is a public health
problem. Antibiotic resistance is caused by the mecA
gene located in the SCCmec chromosome
cassette. The SCCmec type differentiates
between hospital- and community-acquired MRSA and predicts possible
antibiotic resistance genes other than beta-lactams. Few studies have been
done in Venezuela, and in the state of Aragua it is the first research to be
carried out in four hospitals.
Main findings:
The finding was the high frequency of MRSA with SCCmec
I of hospital origin.
Implications:
The research will contribute to establish measures for epidemiological
control and the use of antibiotic therapy in four health centers in the state
of Aragua, Venezuela. |
MATERIALS AND METHODS
Study design
Cross-sectional descriptive study conducted between
January and August 2015 in patients who attended four health centers in the
state of Aragua, Venezuela, called A, B, C and D in this research. Health
center A is a private hospital with 72 beds, outpatient consultation and
hospitalization. Health center B is a public care facility for patients with
diabetic foot complications, which receives an average of 70 people per day and
has no hospitalization. Health center C is a public, preventive care facility
for adult and pediatric patients with a capacity for 2000 people, while health
center D is a public facility and the largest in the region, with a capacity for 400,000 people
and 551 hospitalization beds.
Staphylococcus strains were isolated
from samples of
skin, soft tissue, catheters, auricular, ocular, and respiratory secretions, as
well as other infections. It was identified whether the samples were from
inpatients or outpatients.
These samples were inoculated into blood agar plates and incubated at
35±2 °C in aerobic conditions for 16-18 hours. The standard procedure
described in the literature was used for bacterial identification (16).
Finally, of a total of 404 staphylococcus-positive cultures, S. aureus
was isolated in 324. The strains were preserved at -20 °C in glycerol
until the time of the study.
Antimicrobial susceptibility test
DNA extraction
It
was performed on a MRSA pure culture on blood agar after 18-24 hours of
incubation. A suspension was prepared in an Eppendorf tube by taking 1 to 5
colonies of the microorganism and placing them in 50 µL of sterile
distilled water and then boiled at 99 °C for 10 min. Finally, it was
centrifuged at 30,000 g for 1 min and the supernatant was transferred to a new
Eppendorf tube. The concentrated DNA was preserved at -20 °C until the
time of testing (13).
Detection of the mecA
gene and SCCmec genotypes of MRSA strains
The
multiple PCR test was performed to identify the SCCmec
cassette type and the conditions for amplification according to the methodology
previously described by Zhang et al (13). This methodology
consisted in using 9 pairs of primers, including the specific primers for SCCmec I, II, III, IVa, IVb, IVc, IVd
and V types and subtypes and the primers for the mecA
gene. Eight different loci were amplified based on the sequences presented in
Table 1. The following conditions were used for the PCR reaction: for the
Master Mix, 50 mM KCl,
20 mM Tris-HCl (pH 8.4);
2.5 mM MgCl2, 0.2 mM
of each deoxynucleotide triphosphate (dATP, dUTP, dGTP
and dCTP). The primers concentrations are shown in
Table 1. Additionally, a Go Taq Flexi DNA Polymerase® unit (Promega Corp., USA)
was used.
Table
1. Sequences of the primers that amplify each of the loci of the
Staphylococcal Chromosome Cassette (SCCmec)
Source: Zhang et
al (13)
For controlling quality of the molecular typification tests, the S.
aureus ATCC 259233 (methicillin-sensitive) strain was used as a negative
control and the S. aureus ATCC 43300 (methicillin-resistant) strain was used as a positive control for the mecA gene. The amplification product was subject to
electrophoretic migration in 2% agarose gels at 100 v for 30 min. A 100 bp
molecular size marker (New England Biolabs, Inc) was used. Finally, the length
of the amplicon was compared with the molecular size values recorded in Table 1
for the mecA gene and the SCCmec
types and subtypes.
Statistical analysis
The
provenance data of MRSA strains was collected in a Windows XP Excel 2007
database. Statistical analysis of the data was performed with EpiInfo 3.5.1. Descriptive analyses were performed using
frequencies and percentages. The Chi-square test with a significance level of p
< 0.05 was used to identify differences regarding some characteristics in
the strain’s origin.
Ethical aspects
This
study was evaluated and approved by the Bioethics Committee of the Teaching and
Research Department of the Servicio Autónomo Hospital Central de Maracay. Additionally,
patients who participated in the study signed an informed consent form.
RESULTS
During
the study period a total of 404 staphylococcus-positive cultures from the four
health centers in the state of Aragua were analyzed, 80 strains (19.8%) were
coagulase-negative staphylococci (CSN) and 324 (80.2%) were S. aureus,
of the strains analyzed 81 (25%) were MRSA.
Detection of the mecA gene showed that
from the 81 MRSA isolates, 55 (67.9%) amplified the mecA
gene and 26 (32.1%) did not, with a confidence interval of 56.6% to 77.8% and a
95% confidence level. Among the 55 isolates that tested positive for the mecA gene, only 24 (43.6%) amplified some type of SCCmec, while in 31 isolates (56.4%) no
amplification was obtained with confidence intervals between 30.3%-57.7% and
42.3%-68.7%, respectively (Figure 1A, B and C).
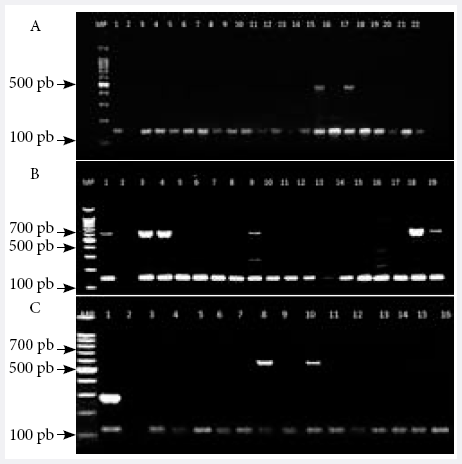
Figure 1. Electrophoresis in 2% agarose gels of the PCR amplification products of SCCmec genotypes and the mecA gene. A) MP: 100 bp molecular weight marker; lane 1: positive control; lane 2: negative control; lanes 3 to 22 MRSA isolates where 147 bp band corresponding to the mecA gene is observed; lanes 15 and 17 have additional 493 bp band corresponding to SCCmec IVb. B) MP: molecular weight marker of 100 bp; lane 1: positive control; lane 2: negative control; lanes 3 to 19 MRSA isolates where a 147 bp band corresponding to the mecA gene is observed; the 881 bp band is SCCmec IVd, 613 bp is SCCmec I, 395 bp corresponds to SCCmec II, 325 bp is SCCmec V and 200 bp IVc; lanes 1, 3, 4, 9, 18 and 19: SCCmec I genotype strains; lanes 5 to 8, 10 to 17: there was no amplification with the SCCmec included in the study C) MP: 100 bp molecular weight marker; lane 1: positive control; lane 2: negative control; lanes 3 to 14 MRSA isolates showing 147 bp band corresponding to the mecA gene; lanes 8 and 10: SCCmec I genotypes strains.
From the 24 MRSA strains that amplified SCCmec,
the most predominant cassette among the isolates was found to be the SCCmec type I, followed by SCCmec
IV (subtypes IVb and IVd)
and SCCmec III represented by 62.5%, 25% and
12.5%, respectively. SCCmec II and SCCmec V were not found (Table 2).
Table
2. Genotype frequency of the staphylococcal chromosome
cassette and its distribution according to health centers and sample type.
|
Characteristic |
Type of staphylococcal chromosome cassette (SCCmec) |
|
|||||
|
I |
II |
III |
IVb |
IVd |
V |
Total |
|
|
n (%) |
n (%) |
n (%) |
n (%) |
n (%) |
n (%) |
n (%) |
|
|
Number of isolates |
15 (62.5) |
0 (0) |
3 (12.5) |
3 (12.5) |
3 (12.5) |
0 (0) |
24 (100) |
|
Health centers |
|
|
|
|
|
|
|
|
A |
12 (80) |
0 (0) |
3 (20) |
0 (0) |
0 (0) |
0 (0) |
15 (100) |
|
B |
2 (40) |
0 (0) |
0 (0) |
2 (40) |
1 (20) |
0 (0) |
5 (100) |
|
C |
0 (0) |
0 (0) |
0 (0) |
1 (50) |
1 (50) |
0 (0) |
2 (100) |
|
D |
1 (50) |
0 (0) |
0 (0) |
0 (0) |
1 (50) |
0 (0) |
2 (100) |
|
Sample type |
|
|
|
|
|
|
|
|
Hemoculture |
1 (100) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
1 (100) |
|
Skin and soft tissue |
12 (63) |
0 (0) |
2 (10) |
3 (16) |
2 (10) |
0 (0) |
19 (100) |
|
Respiratory |
2 (50) |
0 (0) |
1 (25) |
0 (0) |
1 (25) |
0 (0) |
4 (100) |
The largest number of MRSA strains with some type of SCCmec
amplification was found in health center A (15 strains), where SCCmec I was found to be predominant (80%), followed
by SCCmec III (20%). In the case of health center
B, a total of 5 strains amplified SCCmec,
where it was found that three of them (60%) turned out to be SCCmec IV (subtypes IVb
and IVd) and 40%, SCCmec
I. In health center C, two MRSA strains were obtained that amplified SCCmec IV, one amplified subtype IVb
and the other subtype IVd. In health center D, only
two MRSA isolates amplified SCCmec, one of
which was SCCmec I and the other SCCmec IVd. A correlation was
found between the genotype isolated and the health center (p = 0.032) (Table
2).
Of the 24 strains analyzed, 19 were isolated in skin and soft tissues, 4
in secretions from respiratory infections and 1 in blood culture. Of the 19
isolates from skin and soft tissues, SCCmec I
was the most predominant (63.2%), followed by SCCmec
IV (26.3%), where 15.8% were subtype IVb, and 10.5%
subtype IVd. Four strains were obtained in
respiratory samples that amplified SCCmec,
from which two (50%) carried SCCmec I, while
one was SCCmec III and one was SCCmec IVd. No
relationship was found between genotype and type of infection (p = 0.870)
(Tabla
2).
DISCUSSION
In
this study, 25% of MRSA data was similar to the data obtained by Dorante et al (18), who found that out of
117 S. aureus isolates in a hospital in the state of Aragua, 24.7% were
MRSA. Likewise, Chavez et al (19) reported that, in a
hospital in Medellín (Colombia), out of 35 isolates, 28.6% were MRSA. However,
Guillen et al (20) found in Paraguay that, from 77 strains,
18.7% were MRSA, slightly lower than the percentage reported in this study.
On the other hand, detection of MRSA using mecA
gene identification by PCR, showed discrepancies with the results obtained with
the use of oxacillin and cefoxitin discs because from the 81 MRSA isolates,
only 55 amplified the mecA gene. Other studies
have reported similar results, such as Acuña et al
(14), who observed that from 21 MRSA strains, only 19 amplified the mecA gene. In contrast, the research by Chavez et
al (19) and Guillen et al (20) reported that
all of the MRSA strains they studied possessed the mecA
gene. In this study, the strains that did not amplify the mecA
gene had their identification and their phenotypic methicillin resistance
profiles confirmed.
According to the results obtained, it is possible that resistance is
related to some mechanism other than the expression of PBP2a. One of them may
be the hyperproduction of β-lactamases by S. aureus strains, known as
BORSA (Borderline oxacillin-resistant S. aureus) (21). In
addition, it would be possible to imply that the lack of amplification of the mecA gene may be due to the fact that the strain
carries the mecC gene, which is not
detectable by conventional methods, and is responsible for 2% of MRSA
infections in humans. The mecC gene is 70%
homologous to the mccA gene and synthesizes a
transpeptidase that is 60% homologous with PBP2a (22).
Regarding the typification of SCCmec, 24 strains amplified some type of SCCmec, with SCCmec
I being the most predominant, followed by SCCmec
IV (subtypes IVb and IVd)
and SCCmec III in a smaller proportion. Distribution
of SCCmec genotypes was different in health
centers A, B, C and D, health center A had the highest number of strains found,
with a clear predominance of SCCmec I and, in
less frequency, type III, which confirms its in-hospital origin and
demonstrates that those isolates carrying type III, must possess resistance to
a wide variety of antibiotics other than β-lactams (5,8). Center B
occupied the second position according to the total number of isolates, with
predominance of SCCmec type IV (subtypes IVb and IVd), followed by type I.
The presence of SCCmec IV in health center B,
an outpatient center for diabetic foot complications, seems to indicate its origin
in the community (5, 8). However, in the same center, the finding of
SCCmec I is related to MRSA-H strains (5,8), which could predict the possible dissemination of
strains acquired in the hospital environment at this health center.
In addition, SCCmec type IV was mainly
found in health centers C and D. However, the number of MRSA strains associated
to some type of SCCmec was low, which could be
explained by the low resources from most bacteriology laboratories and this
affects detection and identification of MRSA strains. The low number of
detected MRSA strains and SCCmec is considered
a limitation to interpret the results of this study. Therefore, further
research will be necessary to deepen the data about distribution of SCCmec in both health centers, being health center D
the largest and most important one in the region.
Results related to the high frequency of SCCmec
I are similar to those reported in other regions of Venezuela and in other
cities of Latin America. In fact, Acuña et al (14)
found in a hospital in Cumaná (Venezuela) the
predominance of SCCmec I (14 of 19 MRSA
isolates), followed by SCCmec IV (3 of 19 MRSA
strains). In Valdivia, Chile, SCCmec I was
identified, followed by SCCmec IV (23).
The cited investigations also coincide with the predominance of SCCmec I found in health center A. On the other
hand, the presence of SCCmec IV in health
centers B and C, coincides with that proposed by Romero et al (24)
and Castellano et al (25) in hospitals in the state of Zulia,
as well as that referred to by Sanchez et al (26) in
hospitals in Medellín (Colombia). A study carried out in a hospital in Cali
(Colombia) (19) reported that 26.6% of MRSA strains carried SCCmec II, unlike what was observed in this study in
the four health centers investigated. This result could be due to differences
in the predominance and distribution of SCCmec
between hospitals and the geographical area of reference. In fact, the studies
published to date in other regions of Venezuela have not identified SCCmec II (14,15,24,25).
Identifying the presence of SCCmec I in
health centers in the state of Aragua could improve therapeutic options for the
treatment of MRSA infections (5,8).
However, the presence of SCCmec IV also gains
relevance, because of its exclusive resistance to beta-lactam antibiotics and
because it is related to MRSA-C (5,8). MRSA-C isolates with SCCmec IV, and to a lesser extent type V, also carry
the genes for Panton-Valentine Leukocidin toxin (PVL)
(5), while MRSA strains acquired in the hospital environment, have SCCmec II or III, and in very few cases LPV has been
found (5,8). That is why it has been proposed that the identification
of PVL in hospital strains carrying SCCmec IV
allows to corroborate its origin and to clarify the epidemiological panorama (5,8).
In relation to the infection localization, most of the typified strains
came from skin and soft tissues, with predominance of SCCmec
I, confirming the hospital origin of the infections and orienting antibiotic
therapy, since type I is a carrier of resistance to beta-lactam antibiotics (5,8). Results from this study differ from those found
by Romero et al. (24), who found a high percentage of SCCmec IV isolates in skin and soft tissue samples.
From the 55 MRSA isolates that amplified the mecA
gene, 56.4% did not amplify the SCCmec, and it
is possible to assume that primers were used to detect SCCmec
I to V and their subtypes, which was proposed by Zhang et al (13).
This was another limitation for this study. The possible existence of other SCCmecs would indicate the presence of other
genotypes with new antibiotic resistance determinants.
In conclusion, the identification of MRSA by SCCmec
typification showed evidence of the predominance of SCCmec I and III related to MRSA acquired in the
hospital environment and SCCmec type IV
associated to the community. It was shown that there is a correlation between
the isolated genotype and the health center. In skin and soft tissue samples, SCCmec I predominated; however, no correlation was
found between SCCmec and the type of
infection.
It is recommended to carry out prospective studies regarding the
detection of SCCmec, including the main health
centers in the region, the use of alternative methods to verify methicillin
resistance, and the introduction of new primers to identify the existence of
other SCCmec. In addition, it is recommended
to include the assessment of hyperproduction of beta-lactamases and the
determination of minimum inhibitory concentrations of oxacillin for those MRSA
strains that do not amplify the mecA gene.
REFERENCES
1. Organización Mundial de la Salud. 2018. Comunicado de Prensa. Disponible en: https://www.who.int/es/news-room/fact-sheets/detail/resistencia-a-los-antimicrobianos.
2. Organización Mundial de la Salud. 2017. Comunicado de Prensa. Disponible en: https://www.who.int/es/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed.
3. Hiramatsu K, Kuroda M, Ito T. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends. Microbiol. 2001;9(10):486-493. doi: https://doi.org/10.1016/s0966-842x(01)02175-8.
4. Cortés J, Gómez C, Cuervo S, Leal A. Implicaciones en salud Pública de Staphylococcus aureus meticilino resistente adquirido en la comunidad en Bogotá, Colombia. Rev. Salud Pública. 2007;9(3): 448–454. Disponible en: https://www.scielosp.org/pdf/rsap/2007.v9n3/448-454/es.
5. Ito T, Katayama Y, Asada K, Namiko T, Kanae T, Chuntima T et al. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin- resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2001;45(5):1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001.
6. Oliveira D, Tomasz A, De Lancastre H. The evolution and pandemic clones of methicillin-resistant Staphylococcus aureus: Identification of two ancestral genetics brack grounds and the associated mec elements. Microb. Drug. Resist. 2001;7(4):349-361. doi: 10.1089/10766290152773365.
7. Liu J, Chen D, Peters B M, Li L, Li B, Xu Z, et al. Staphylococcal chromosomal cassettes mec (SCCmec): A mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb Pathog. 2016;101:56-67. doi: https://doi.org/10.1016/j.micpath.2016.10.028.
8. Zhang K, McClure JA, Elsayed S, Conly JM. Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2009; 53(2):531–540. 10.1128/AAC.01118-08.
9. Canoa M, Domínguez M, Ezpeletac C, Padillad B, Arellano E y Martínez L. Cultivos de vigilancia epidemiológica de bacterias resistentes a los antimicrobianos de interés nosocomial. Enferm Infecc Microbiol Clin. 2008;26(4):220-229. doi: 10.1016/S0213-005X(08)72694-6.
10. Pérez N, Pavas N , Rodríguez E. Resistencia de Staphylococcus aureus a los antibióticos en un hospital de la Orinoquia colombiana. Asociación Colombiana de Infectología. Rev Infect. 2010;14(3):167-173. doi: 10.1016/S0123-9392(10)70108-9.
11. Cavalcante F, Schuenck R, Caboclo R, Ferreira D de C, Nouér S, Dos Santos K. Tetra cycline and trimethoprim/sulfamethoxazole at clinical laboratory: can they help to characterize Staphy lococcus aureus carrying different SCCmec types?. Rev Soc Bras Med Trop. 2013;46(1):100-102. doi: 10.1590/0037-868216062013.
12. Faria N, Carrico J, Oliveira D, Ramírez M, De Lencastre H. Analysis of typing methods for epidemiological surveillance of both methicillin–resistant and methicillin susceptible Staphylococcus aureus strains. J. Clin. Microbiol. 2008; 46(1):136–144. doi: 10.1128/JCM.01684-07.
13. Zhang K, McClure J, Elsayed S, Louie T. Novel Multiplex PCR Assay for Characterization and Concomiitant Subtyping of Staphylococcal Cassette Chromosome mec Types I to V in Methicillin-Resistant Staphylococcus aureus. J. of Clin. Microbiol. 2005;43(10):5026-5033. doi: 10.1128/JCM.43.10.5026-5033.2005.
14. Acuña S, Sánchez E y Patiño L. Tipificación de la meticilino resistencia en cepas de Staphylococcus spp. Hospital Universitario de Alcalá, Cumana, Estado Sucre, Venezuela. Rev. Soc. Ven. Microbiol. Caracas. 2014;34(1):1-8. Disponible en: http://ve.scielo.org/pdf/rsvm/v34n1/art03.pdf.
15. González M, Cavazza M, Perozo A. Tipo de cassette cromosómico estafilocócico en cepas clínicas de Staphylococcus aureus resistentes a meticilina. Kasmera. 2014;42(2):116-130. Disponible en: http://ve.scielo.org/pdf/km/v42n2/art04.pdf.
16. Koneman E., Allen S., Janda, W., Schreckenberg P. y Winn W. Diagnóstico Microbiológico. (5ª Ed.). Madrid: Panamericana; 2008.
17. Instituto de Estándares Clínicos y de Laboratorio. Normas de funcionamiento de los antimicrobianos, Las pruebas de susceptibilidad de disco. Undécima edición. Estados Unidos. Estados Unidos; 2012.
18. Dorante V, Hurtado E, Martínez B, Méndez MV. Frecuencia de Staphylococcus aureus meticilino resistente en pacientes que asisten al laboratorio de microbiología del hospital "Los Samanes" estado Aragua. Odous Científica. 2013;14(1):29-36. Disponible en: http://servicio.bc.uc.edu.ve/odontologia/revista/vol14-n1/art04.pdf.
19. Chávez M, Martínez A, Esparza M. Caracterización de Staphylococcus aureus obtenido del ambiente hospitalario y del personal de salud en un Hospital de la Ciudad de Cali. Rev Biosalud. 2017;16(2):22-33. doi: 10.17151/biosa.2017.16.2.3.
20. Guillén R, Carpinelli L, Rodríguez F, Castro H, Quiñónez B, Campuzano A. Staphylococcus aureus adquiridos en la comunidad: caracterización clínica, fenotípica y genotípica de aislados en niños paraguayos. Rev Chilena Infectol. 2016;33(6): 609-618. doi: 10.4067/S0716-10182016000600002.
21. Velásquez-Meza, M. Surgimiento y Diseminación de Staphylococcus aureus meticilino resistente. Salud Pública Mex. 2005;47:381-387. Disponible en: http://www.scielo.org.mx/pdf/spm/v47n5/28384.pdf.
22. Ito T, Hiramatsu K, Tomasz A, de Lencastre H, Perreten V, Holden M T, et al. Guidelines for reporting novel mecA gene homologues. Antimicrob Agents Chemother. 2012;56(10):4997-4999. doi: 10.1128/AAC.01199-12.
23. Medina G, Egea A L, Otth C, Otth L, Fernández H, Bocco J L, et al. Molecular epidemiology of hospital-onset methicillin-resistant Staphylococcus aureus infections in Southern Chile. Eur J Clin Microbiol Dis. 2013;32(12):1533-1540. doi: https://doi.org/10.1007/s10096-013-1907-8.
24. Romero A, Castellano M, Perozo M, Armindo J, Rincón G, Zabala D. Detección de cassette cromosómico en cepas de Staphylococcus aureus resistente a meticilina aisladas en un hospital universitario de la ciudad de Maracaibo. Kasmera. 2018; 46(1):40-51. Disponible en: https://produccioncientificaluz.org/index.php/kasmera/article/view/24652/pdf.
25. Castellano M, Cavazza J, Porro M, Perozo M, Armindo J. Tipo de cassette cromosómico estafilocócico en cepas clínicas de Staphylococcus aureus resistentes a meticilina. Kasmera. 2014;42(2):116-130. Disponible en: https://produccioncientificaluz.org/index.php/kasmera/article/view/19528/19492.
26. Sánchez M, Hernández O, Velásquez L, Rivas D, Marín A, González L, et al. Caracterización del gen mecA de Staphylococcus aureus resistentes a meticilina aislados de tres grupos poblacionales de la ciudad de Medellín. Infectio. 2013;17(2):66-72. doi: 10.1016/S0123-9392(13)70165-6.
Citation: Bastidas B, Méndez MV, Vásquez Y, Requena D. Typification of the staphylococcal chromosome cassette of methicillin-resistant Staphylococcus aureus in the state of Aragua, Venezuela. Rev Peru Med Exp Salud Publica. 2020;37(2):239-45. doi: https://doi. org/10.17843/rpmesp.2020.372.4652
Correspondence to: María V. Méndez; mvmendezster@gmail.com
Funding sources: The study was funded by the Center for Scientific and Humanistic Development of the University of Carabobo-Venezuela (CDCHUC).
Authorship contributions: BB, MVM, YV and DR have participated in the conception of the article, data collection, writing and approval of the final version.
Conflicts of interest: The authors declare that there is no conflict of interest regarding the publication of this article.
Received: 07/07/2019
Approved: 08/04/2020
Online: 15/06/2020