ORIGINAL ARTICLE
Mutations conferring resistance to first-line anti-tuberculosis drugs in Peru: a systematic review of the literature
Aiko Vigo1,a, Lely Solari1,b, David Santos1,a, Zully M. Puyén.1,a,c
1 Instituto Nacional de Salud, Lima, Perú.
a Licenciate in Biology; b Infectologist, PhD in Health Sciences; c doctorate in Microbiology.
ABSTRACT
Objective. To systematize available information regarding mutations that confer resistance to first-line anti-tuberculosis drugs. Materials and Methods. A systematic review of the scientific literature was conducted to identify articles that reported mutations conferring resistance to first-line anti-tuberculosis drugs. This search emphasized resistance to isoniazid and rifampicin drugs in M. tuberculosis strains of Peruvian patients. The search was performed on PubMed and LILACS (Latin American and Caribbean Health Sciences Literature). Results. Fourteen (14) articles were included, of which three reported mutations associated with resistance to isoniazid, six to rifampicin, eight to pyrazinamide and one to ethambutol. All mutations to isoniazid or rifampicin were identified directly or indirectly by the molecular diagnostic test GenoType MTBDRplus® v2.0. The greatest variability of mutations was determined in resistance to pyrazinamide. Conclusions. There is a great variability of mutations associated with resistance to anti-tuberculosis drugs that have been reported in Peru, and they are systematized in this report. These mutations must be taken into account for the development of diagnostic devices or selection of diagnostic tests to be applied in our country.
Keywords: Mycobacterium tuberculosis; Drug Resistance; Genotype; Mutation (source: MeSH NLM).
INTRODUCTION
Drug-resistant tuberculosis (TB) is a major global public health problem. In 2018, the World Health Organization (WHO) estimated that ten million people developed the disease. Of these cases, 500,000 had rifampicin-resistant tuberculosis, of which 78% was multidrug-resistant tuberculosis (MDR-TB); that is, they had simultaneous resistance to isoniazid and rifampicin (1). In addition, it was determined that 7.3% of the diagnosed MDR-TB cases were extensively drug-resistant tuberculosis (XDR-TB) cases; that is, they were additionally resistant to at least one fluoroquinolone (levofloxacin, ciprofloxacin, or moxifloxacin) and a second-line aminoglycoside (capreomycin, amikacin, kanamycin). Peru is no stranger to MDR-TB and XDR-TB epidemics, being among the 30 countries with the highest MDR-TB burden in the world (1).and reporting around 3000 prevalent cases of MDR-TB annually (2).
Resistance to anti-tuberculosis drugs is determined by mutations in the M. tuberculosis genome, which occur spontaneously or induced by selective drug pressure (3). Patients who take inadequate therapeutic schemes or who are not very adherent can generate, in the presence of subtherapeutic concentrations of the drugs, proliferation of the strains that present these genetic resistance mutations (3). These strains can then be transmitted by air to other people, who develop drug-resistant tuberculosis without having previously received any therapeutic regimen.
Mutations present in the coding gene of the "RNA polymerase subunit β" (rpoB gene) are now known to confer resistance to rifampicin. Likewise, mutations present in the "catalase-peroxidase" gene (katG gene) and in the promoter region of the "enoyl-ACP reductase" gene (inhA gene) are associated with resistance to isoniazid (4). On the other hand, molecular diagnostic devices used in our health system, such as the GenoType MTBDRplus® v2.0 (Hain Lifescience) and Xpert® MTB/ RIF (Cepheid, USA) detect resistance to the abovementioned drugs. There are also mutations in other genes that have been identified as responsible for resistance to other anti-tuberculosis drugs such as pyrazinamide (pncA), aminoglycosides (rrs), fluoroquinolones (gyrA), among others.
The presence and/or absence of mutations can vary significantly from one country to another, which could mean that the diagnostic devices that identify the most frequent mutations in one country does not have the same diagnostic performance in another (3). Therefore, knowledge of mutations existing in strains of M. tuberculosis which circulate at the national level can help develop new diagnostic devices that improve resistance detection at a local level (5).
The objective of this study is to review the available information regarding mutations that confer resistance to firstline anti-tuberculosis drugs, with emphasis on isoniazid and rifampicin, in publications that describe strains of M. tuberculosis from Peruvian patients.

MATERIALS AND METHODS
A systematic review of the scientific literature related to the molecular characterization of mutations existing in M. tuberculosis strains with resistance to first-line anti-tuberculosis drugs in Peru was performed, according to PRISMA guidelines (6). The study focused on the analysis of resistance to isoniazid and rifampicin, as well as strains of M. tuberculosis isolated from peruvian patients.
INCLUSION AND EXCLUSION CRITERIA
The inclusion criteria for the publications considered in the study were first, language (articles in english, spanish, french, and portuguese); second, study design (case reports or series, observational or experimental studies); third, that they are articles with explicit information on the nucleotide sequence of the mutation conferring resistance to first-line drugs, mainly isoniazid and rifampicin (independently of any additional tests they may have done, including non-commercial and commercial tests); fourth, that they include strains from Peruvian patients; and finally, that they have been published in the last ten years, with the aim of obtaining a better reflection of the diversity of mutations conferring resistance today.
Articles that only included phenotypic tests for the diagnosis of resistance, strains of patients from countries other than Peru, and articles aimed at studying the genetic lineage of mycobacteria, but not their resistance to anti-tuberculosis drugs, were excluded.
BIBLIOGRAPHIC SEARCH STRATEGY
Bibliographic databases searched to identify studies to be included were: PubMed, the Library of Congress, and LILACS (Latin American and Caribbean Literature in Health Sciences).
For the PubMed search, the following strategy was used: "(polymorphism OR genotype OR variant OR mutation OR allele) AND Peru AND tuberculosis", words that were selected based on previous systematic reviews about mutations (7,8). The LILACS search used the following strategy: «mutaciones» y «tuberculosis» y «Peru» ("mutations" and "tuberculosis" and "Peru").
In the case of multiple publications on the same study, the study with the largest sample or first to be published was included.
STUDY SELECTION AND DATA COLLECTION
AV and LS independently selected the studies to be included. The list of included and excluded studies was then reviewed. Where there was conflict, it was discussed, and consensus was reached. References and articles related to the initially identified publications were also reviewed.
Once the studies to be included were defined, information about the characteristics of the publication (author, journal, year), the study (type of design, characteristics of patient), and the strains (evaluated drugs, evaluated genes and existing mutations) was extracted into tables previously designed for data collection. Finally, "summary tables" were elaborated with the collected information.
The quality of the publications was not formally evaluated since they are often specific reports of mutations and the medical literature for the reporting of this type of publications is heterogenous (initiatives to homogenize the reports of publications about mutations in M. tuberculosis are just emerging). For that reason, it was decided not to carry out a risk of bias assessment in the studies.
The main aspect to report was the nucleotide sequences of specific mutations associated with resistance to first-line anti-tuberculosis drugs, particularly isoniazid and rifampicin. Finally, once the information on the mutations found was extracted, they were graded as minimal, moderate and high confidence for association with resistance according to the WHO’s technical guide on "The use of next-generation sequencing technologies for the detection of mutations associated with drug resistance in Mycobacterium tuberculosis complex" (9). The results are reported according to the anti-tuberculosis drug analyzed.
It is worth mentioning that the protocol of this review was not registered and, since it was a purely descriptive topic, the respective PICO question was not structured.
RESULTS
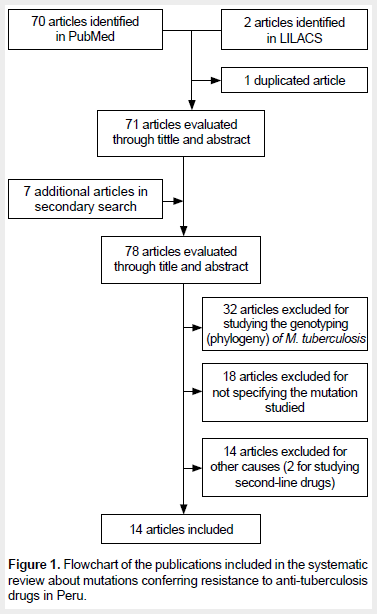
Seventy (70) articles were retrieved from PubMed and two from LILACS, of which one was duplicated. In addition, other seven articles were obtained from the references in the selected articles. Sixty-four (64) articles were excluded, and, finally, 14 studies met our inclusion/exclusion criteria and were admitted for review. Figure 1 shows the selection flowchart for the included studies. The studies of Moradigaravan (10) and Barletta (11) were excluded for studying second-line drugs (para-amino salicylic acid, and quinolones and aminoglycosides, respectively).
The main characteristics of these studies are shown in Table 1. Of the 14 articles, 13 were published in English. Nine articles focused on the evaluation of mutations that conferred resistance to a single anti-tuberculosis drug and seven evaluated more than one drug at a time. Characteristics such as population type and specimens are also described in Table 1.
First-line anti-tuberculosis drugs (isoniazid and rifampicin) are discussed in Tables 2 and 3, respectively.
In addition, information concerning pyrazinamide is included in the supplementary material. Studies evaluating several drugs simultaneously were included in all the respective tables.
Of the three studies that reported resistance to isoniazid, two evaluated the inhA and katG genes, and one evaluated the katG and kasA genes.
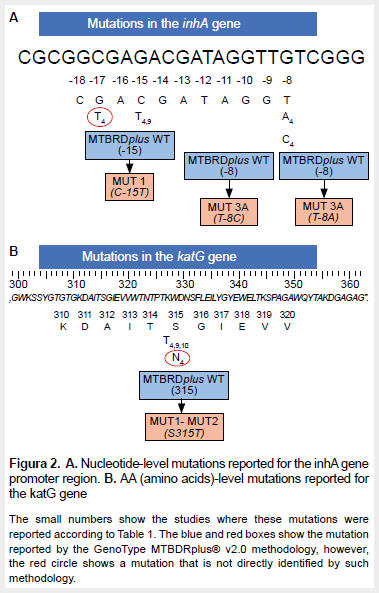
Of the seven mutations reported in these studies, two had a high likelihood of association with resistance and one had a moderate likelihood of association with resistance (26). The most frequent mutation was reported in codon 315 of the katG gene. The location of these mutations in the inhA and katG genes, as well as the gene region evaluated by the commercial test for resistance diagnosis (GenoType MTBDRplus® v2.0), is shown in Figure 2.
Of the six studies that reported mutations associated with resistance to rifampicin, all showed mutations in the rpoB gene, and it could be seen that Linger's study (15) identified the most mutations. The studies of Maha, Gygli, and Galarza (12, 13, 20) mostly presented mutations with a high likelihood of association with resistance. The two most frequent mutations, D516V and H526Y, had a high association with resistance (Table 3, Figure 3).
With regards to mutations in the pncA gene associated with resistance to pyrazinamide, eight studies that reported a high number of mutations were identified. In addition, it could be seen that the mutations showed a high association with resistance to this drug. The most frequently found mutation was H51R, which is highly associated with resistance. Mutations conferring resistance to pyrazinamide already classified by WHO and those pending classification are reported in the supplemental material.
With respect to the other first-line anti-tuberculosis drugs, one study (12) evaluating the rpsL gene for resistance to streptomycin was observed. This study used Whole Genome Sequencing and 7H10 and MGIT BACTEC 960 as phenotypic method, finding the K43R mutation, which is highly associated with resistance. Likewise, Galarza also used Whole Genome Sequencing to evaluate resistance to ethambutol through the embB gene, finding the Y319S mutation, which does not confer resistance (21).
DISCUSSION
This systematic review identified 14 studies reporting mutations in the genome of M. tuberculosis in genes associated with resistance to first-line anti-tuberculosis drugs. In total, seven mutations in relation to resistance to isoniazid were reported (two of them with high and one with moderate confidence for association with resistance). For rifampicin, of the 31 mutations found, 21 have already been confirmed as associated with resistance (15 with high, four with moderate, and two with minimal confidence for association with resistance). Additionally, only one mutation associated with resistance to ethambutol, one to streptomycin, and 48 to pyrazinamide —a drug on which a significant number of research studies have been conducted— were found. In addition to the objectives of the study, we were able to identify three mutations associated with aminoglycoside resistance and five with quinolone resistance.
This denotes an important effort from the tuberculosis research groups present in our country, many of which have been very active during the last decades, producing studies to deepen the knowledge of the epidemiology, clinical and microbiology of tuberculosis in Peru. In recent years, the branch that has gained the most relevance is molecular biology, which works to build the foundations not only for new diagnostic methodologies by detecting mycobacterial genetic material, but also for the development of new drugs (26).
It is important to point out that, as part of the control of drug-resistance in tuberculosis, the priority must be the identification of resistance to the two pillars of treatment: isoniazid and rifampicin, which is why these drugs were the focus of the study. For detection of rifampicin resistance, commercially available and recommended tests are: GenoType MTBDRplus® v2.0 line probe assay and Xpert® MTB/RIF. Both tests detect mutations in the "Rifampicin Resistance-Determining Region" (RRDR) using DNA probes that bind specifically to that region. A comparative study carried out in 2014 between GenoType MTBDRplus® v2.0 and Xpert® MTB/RIF determined the superiority of the former for the detection of rifampicin mono-resistant M. tuberculosis in terms of sensitivity (27). Furthermore, isoniazid resistance can only be detected using the GenoType MTBDRplus® v2.0 test taking into consideration two genes associated with resistance: the katG gene and the promoter region of inhA gene. The Xpert® MTB/RIF test does not detect this resistance.
At the katG gene level, codon 315 was the only position that evidenced the appearance of two mutations highly associated with isoniazid resistance: S315T and S315N (9,15). When evaluating the mutations detected by GenoType MTBDRplus® v2.0 (Figure 2) we can see that only the S315T mutation is directly identified, whereas the S315N mutation (15) is indirectly identified. Similarly, when comparing the information collected on mutations found in the inhA gene by the GenoType MTBDRplus® v2.0 test, the mutation present in the g-17t nucleotide position is indirectly identified. S315N and g-17t mutations in the katG and inhA genes, respectively, are not detected directly by the GenoType MTBDRplus® v2.0 test, but indirectly by the absence of some wild type band. On the other hand, the G269S mutation of the kasA gene is not analyzed by this molecular test.
With respect to the mutations reported in the rpoB gene, in general, all mutations can be detected by molecular tests GenoType MTBDRplus® v2.0 and Xpert® MTB/RIF. In addition to the mutations usually associated with resistance, mutations L511R, D516E, N518H, Q510H, S512R, L524S, H526Q and S531C were identified (15, 20), which are not reported in the WHO technical guide (9). These mutations D516E, D516G, D516Y, del507, H526C, H526L, H526N, H526P, H526Q, H526R, L511P, L511R, L524S, L533P, M515I, N516H, Q510H, Q513K, Q513L, Q513P, S512T, S512R, S522L, S531C, S531F, S531Q, S531W are not defined in the GenoType MTBDRplus® v2.0 mutation bands; however, they are detected indirectly by this methodology, due to the absence of wild type bands covering the respective codons.
Although some mutations are not directly determined by the GenoType MTBDRplus® v2.0 test for both isoniazid and rifampicin, they will be reported as inferred resistance due to the absence of wild type bands. Something similar occurs with the Xpert® MTB/RIF test, as it works with wild probes and does not identify specific mutations (28). The Xpert® MTB/RIF hybridization probes A, B, C, D, and E indirectly identify all rifampicin mutations reported in this systematic review (29,30).
Another important finding is the large number of mutations conferring resistance to pyrazinamide (Annex 3). Microbiological detection of resistance to this drug has always been a challenge (26), and, to date, the most widely used method is Wayne’s (31). The fact that there are a large number of mutations associated with pyrazinamide resistance makes it difficult to develop molecular methods of resistance detection.
In 2018, the use of whole genome sequencing for the detection of resistance-associated mutations in M. tuberculosis was recommended by WHO for implementation in national reference laboratories (9). Because of this, the use of whole genome sequencing of M. tuberculosis—under usual or routine conditions—will provide more complete information about the significance of mutations for the identification of mycobacterial species, detection of anti-tuberculosis drug resistance, and the dynamics of microorganisms’ transmission (32, 33).
As part of the strengths of our study, it should be noted that there are only few systematic reviews on molecular biology issues, and specifically on resistance to anti-tuberculosis drugs. Previous studies have been published, such as the one by Vásquez-Loarte (34); but they are more focused on the association of polymorphisms to evaluate the phylogenetic origin of M. tuberculosis. However, our study specifically addresses the issue of mutations existing in the strains which circulate in our country. This may be more useful from a practical point of view, since this information gathers data on the mutations that are present in Peruvian strains to date. On the other hand, it can serve as a basis for the development of tests to detect resistance to anti-tuberculosis drugs in our country, considering high-prevalence mutations at the global level, as well as those that are not yet included in commercial diagnostic devices.
We must also point out some aspects of the study that limit its scope. First, because not all publications recorded mutation frequencies, it was not possible to determine the mutations that are most prevalent in our territory. We can only mention that they are present. On the other hand, despite our effort to verify, according to WHO recommendations, whether mutations conferred minimal, moderate or high confidence for association with resistance, or were not associated with resistance, there were numerous mutations that could not be classified. Therefore, the clinical implications of the latter are uncertain for now, until more studies are conducted and more evidence on their impact is generated.
In conclusion, there is a wide variety of mutations associated with resistance to first-line anti-tuberculosis drugs in peruvian patients. For the drugs isoniazid and rifampicin, the mainstays of anti-tuberculosis treatment, the number of resistanceassociated mutations is lower compared to pyrazinamide. A positive aspect is that the vast majority of mutations for both isoniazid and rifampicin are directly or indirectly identified (by the absence of wild type probe hybridization) by the GenoType MTBDRplus® v2.0 and Xpert® MTB/RIF tests. These considerations must be taken into account for the development of diagnostic devices to be used locally in our country.
Authors' Contributions: AV participated in the conception of the study. LS participated in the design of the study. AV, LS, DS, and ZP participated in the analysis and interpretation of the data, the writing of the article, and the approval of the final version. All authors claim responsibility for the study.
Funding: This study was finded by INNOVATE - PERU within the scope of contract - N°353-PNICP-PIAP-2014.
Conflicts of Interest: The authors declare that they have no conflicts of interest.
Supplementary Material: Available in the electronic version of the RPMESP.
REFERENCES
1. World Health Organization. WHO | Global tuberculosis report 2019 [Internet]. WHO. 2019 [citado el 31 de octubre de 2019]. Disponible en: http://www.who.int/tb/publications/global_report/en/
2. Ministerio de Salud. Direccion de Prevencion y Control de la Tuberculosis [Internet]. [citado el 29 de octubre de 2019]. Disponible en: http://www.tuberculosis.minsa.gob.pe/DashboardDPCTB/Dashboard.aspx
3. Loddenkemper R, Sagebiel D, Brendel A. Strategies against multidrug-resistant tuberculosis. European Respiratory Journal. 2002;20(36 suppl):66s-77s.
4. Dahle UR, Sandven P, Heldal E, Mannsaaker T, Caugant DA. Deciphering an Outbreak of Drug-Resistant Mycobacterium tuberculosis. J Clin Microbiol. 2003;41(1):67-72. doi: 10.1128/JCM.41.1.67-72.2003.
5. Boldú J, Cebollero P, Abu J, De Prado A. Tratamiento de la tuberculosis pulmonar. Anales del Sistema Sanitario de Navarra. 2008;30(0):87-98.
6. Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264-9, W64. doi: 10.7326/0003-4819-151-4-200908180-00135.
7. Ramirez-Busby SM, Valafar F. Systematic review of mutations in pyrazinamidase associated with pyrazinamide resistance in Mycobacterium tuberculosis clinical isolates. Antimicrob Agents Chemother. 2015;59(9):5267-77. doi: 10.1128/AAC.00204-15.
8. Whitfield MG, Soeters HM, Warren RM, York T, Sampson SL, Streicher EM, et al. A Global Perspective on Pyrazinamide Resistance: Systematic Review and Meta-Analysis. PLoS One. 2015;10(7):e0133869. doi: 10.1371/journal.pone.0133869.
9. World Health Organization. The use of next-generation sequencing technologies for the detection of mutations associated with drug resistance in Mycobacterium tuberculosis complex: technical guide [Internet]. 2018 [citado el 29 de octubre de 2019]. Disponible en: https://apps.who.int/iris/handle/10665/274443.
10. Moradigaravand D, Grandjean L, Martinez E, Li H, Zheng J, Coronel J, et al. DFRA thyA Double Deletion in para-Aminosalicylic Acid-Resistant Mycobacterium tuberculosis Beijing Strains. Antimicrob Agents Chemother. 2016;60(6):3864-7. doi: 10.1128/AAC.00253-16.
11. Barletta F, Zamudio C, Rigouts L, Seas C. Resistance to second-line anti-tuberculosis drugs among peruvian multidrug resistant Mycobacterium tuberculosis strains. Rev Peru Med Exp Salud Publica. 2014;31(4):676-82.
12. Gygli SM, Keller PM, Ballif M, Blöchliger N, Hömke R, Reinhard M, et al. Whole-Genome Sequencing for Drug Resistance Profile Prediction in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2019;63(4):e02175-18. doi: 10.1128/AAC.02175-18
13. Farhat MR, Sixsmith J, Calderon R, Hicks ND, Fortune SM, Murray M. Rifampicin and rifabutin resistance in 1003 Mycobacterium tuberculosis clinical isolates. J Antimicrob Chemother. 2019;74(6):1477-83. doi: https://doi.org/10.1093/jac/dkz048.
14. Jaramillo L, Tarazona D, Levano KS, Galarza M, Caceres O, Becker M, et al. A rapid identification technique for drug-resistant Mycobacterium tuberculosis isolates using mismatch specific cleavage enzyme. Bioinformation. 2018;14(7):404-7. doi: 10.6026/97320630014404.
15. Linger Y, Knickerbocker C, Sipes D, Golova J, Franke M, Calderon R, et al. Genotyping Multidrug-Resistant Mycobacterium tuberculosis from Primary Sputum and Decontaminated Sediment with an Integrated Microfluidic Amplification Microarray Test. J Clin Microbiol. 2018;56:e01652-17. doi: https://doi.org/10.1128/JCM.01652-17.
16. Rueda D, Bernard C, Capton E, Boudjelloul R, Brossier F, Veziris N, et al. Estimation of pyrazinamidase activity using a cell-free In vitro synthesis of pnca and its association with pyrazinamide susceptibility in Mycobacterium tuberculosis. Int J Mycobacteriol. 2018;7(1):16-25. doi: 10.4103/ijmy. ijmy_187_17.
17. Tessema B, Nabeta P, Valli E, Albertini A, Collantes J, Lan NH, et al. FIND Tuberculosis Strain Bank: a Resource for Researchers and Developers Working on Tests To Detect Mycobacterium tuberculosis and Related Drug Resistance. J Clin Microbiol. 2017;55(4):1066-73. doi: 10.1128/ JCM.01662-16.
18. Sheen P, Requena D, Gushiken E, Gilman RH, Antiparra R, Lucero B, et al. A multiple genome analysis of Mycobacterium tuberculosis reveals specific novel genes and mutations associated with pyrazinamide resistance. BMC Genomics. 2017;18:769. doi: 10.1186/s12864-017-4146-z.
19. Dudley MZ, Sheen P, Gilman RH, Ticona E, Friedland JS, Kirwan DE, et al. Detecting Mutations in the Mycobacterium tuberculosis Pyrazinamidase Gene pncA to Improve Infection Control and Decrease Drug Resistance Rates in Human Immunodeficiency Virus Coinfection. Am J Trop Med Hyg. 2016;95(6):1239-1246. doi: 10.4269/ajtmh.15-0711.
20. Galarza M, Fasabi M, Levano KS, Castillo E, Barreda N, Rodriguez M, et al. High-resolution melting analysis for molecular detection of multidrug resistance tuberculosis in Peruvian isolates. BMC Infect Dis. 2016;16(1):260. doi: https://doi.org/10.1186/s12879-016-1615-y.
21. Galarza M, Tarazona D, Borda V, Agapito JC, Guio H. Evidence of Clonal Expansion in the Genome of a Multidrug-Resistant Mycobacterium tuberculosis Clinical Isolate from Peru. Genome Announc. 2014;2(1):e00089-14. doi: 10.1128/genomeA.00089-14.
22. Sheen P, Lozano K, Gilman RH, Valencia HJ, Loli S, Fuentes P, et al. pncA gene expression and prediction factors on pyrazinamide resistance in Mycobacterium tuberculosis. Tuberculosis (Edinb). 2013;93(5):515-22. doi: 10.1016/j.tube.2013.03.005.
23. Zimic M, Sheen P, Quiliano M, Gutierrez A, Gilman RH. Peruvian and globally reported amino acid substitutions on the Mycobacterium tuberculosis pyrazinamidase suggest a conserved pattern of mutations associated to pyrazinamide resistance. Infect Genet Evol. 2010;10(2):346-9. doi: 10.1016/j.meegid.2009.11.016.
24. Sheen P, Ferrer P, Gilman RH, López-Llano J, Fuentes P, Valencia E, et al. Effect of pyrazinamidase activity on pyrazinamide resistance in Mycobacterium tuberculosis. Tuberculosis (Edinb). 2009;89(2):109-13. doi: 10.1016/j.tube.2009.01.004.
25. Sheen P, Méndez M, Gilman RH, Peña L, Caviedes L, Zimic MJ, et al. Sputum PCR-Single-Strand Conformational Polymorphism Test for Same-Day Detection of Pyrazinamide Resistance in Tuberculosis Patients. J Clin Microbiol. 2009;47(9):2937-43. doi: 10.1128/JCM.01594-08.
26. Miotto P, Tessema B, Tagliani E, Chindelevitch L, Starks AM, Emerson C, et al. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. Eur Respir J. 2017;50(6):1701354. doi: 10.1183/13993003.01354-2017.
27. Rufai SB, Kumar P, Singh A, Prajapati S, Balooni V, Singh S. Comparison of Xpert MTB/RIF with line probe assay for detection of rifampinmonoresistant Mycobacterium tuberculosis. J Clin Microbiol. 2014;52(6):1846-52. doi: 10.1128/JCM.03005-13.
28. Hain Lifescience. GenoType MTBDRs/. Detection of resistance of MTBC complex [Internet]. 2015 [citado el 29 de octubre de 2019]. Disponible en: https://www.hain-lifescience.de/en/products/microbiology/mycobacteria/tuberculosis/genotype-mtbdrsl.html.
29. Organización Panamericana de la Salud/Organización Mundial de la Salud. Manual de Capacitación en GeneXpert [Internet]. OPS/OMS. 2017 [citado el 29 de octubre de 2019]. Disponible en: https://www.paho.org/hq/index.php?option=com_content&view=article&id=12924:manu-al-de-capacitacion-en-genexpert&Itemid=42250&lang=es.
30. Andre E, Goeminne L, Cabibbe A, Beckert P, Kabamba Mukadi B, Mathys V, et al. Consensus numbering system for the rifampicin resistanceassociated rpoB gene mutations in pathogenic mycobacteria. Clin Microbiol Infect. 2017;23(3):167-172. doi: 10.1016/j.cmi.2016.09.006.
31. Leo E, Vásquez L, Asencios L, Quispe N, Gómez L, Lecca L, et al. Determinación de la susceptibilidad de Mycobacterium tuberculosis a la pirazinamida mediante la prueba de la pirazinamidasa, Perú – 1999. Rev Peru Med Exp Salud Publica. 2003;20(2):105-6.
32. Outhred AC, Jelfs P, Suliman B, Hill-Cawthorne GA, Crawford ABH, Marais BJ, et al. Added value of whole-genome sequencing for management of highly drug-resistant TB. J Antimicrob Chemother. 2015;70(4):1198-202. doi: 10.1093/jac/dku508.
33. Quan TP, Bawa Z, Foster D, Walker T, del Ojo Elias C, Rathod P, et al. Evaluation of Whole-Genome Sequencing for Mycobacterial Species Identification and Drug Susceptibility Testing in a Clinical Setting: a Large-Scale Prospective Assessment of Performance against Line Probe Assays and Phenotyping. J Clin Microbiol. 2018;56(2):e01480-17. doi: 10.1128/JCM.01480-17.
34. Vásquez-Loarte T, Trubnykova M, Guio H. Genetic association meta-analysis: a new classification to assess ethnicity using the association of MCP1 -2518 polymorphism and tuberculosis susceptibility as a model. BMC Genet. 2015;16:128. doi: 10.1186/s12863-015-0280-2.
Correspondence to: Aiko Vigo
Address: Defensores del Morro, Chorrillos, Lima, Perú.
Email:
aiko.vt.anvt@gmail.com
Phone: +51924167434
Received: 06/08/2019
Approved: 06/11/2019
Online: 03/12/2019