10.17843/rpmesp.2020.372.4829
BRIEF REPORT
Presence of fimH and afa genes in urinary isolates of extended-spectrum beta-lactamases producing Escherichia coli in Lima, Peru
Jose Matta-Chuquisapon
![]() 1, Medical Technologist
1, Medical Technologist
Esther Valencia-Bazalar
![]() 1, Medical Technologist
1, Medical Technologist
Luis Marocho-Chahuayo
![]() 2, Physician specialized in Infectious and Tropical Diseases
2, Physician specialized in Infectious and Tropical Diseases
Edgar Gonzales-Escalante
![]() 3,4, Medical Technologist, Master in Microbiology
3,4, Medical Technologist, Master in Microbiology
Carlos Raúl Sevilla-Andrade
![]() 3,5, Medical Technologist
3,5, Medical Technologist
1 Universidad
Nacional Mayor de San Marcos, Lima, Perú.
2 Instituto Nacional de Salud del Niño, Lima, Perú.
3 Centro de Investigaciones Tecnológicas, Biomédicas y
Medioambientales (CITBM), Universidad Nacional Mayor de San Marcos, Lima,
Perú.
4 Laboratorio de Resistencia Bacteriana, Facultad de Farmacia y
Bioquímica, Universidad de Buenos Aires, Argentina.
5 Grupo de Investigación en Resistencia a los antimicrobianos
(MICRESIS), Facultad de Medicina, Universidad Nacional Mayor de San Marcos,
Lima, Perú.
This study is part of Matta Chuquisapon J.’ s undergraduate thesis: Frequency of fimH and afa genes in extended-spectrum beta-lactamases producing Escherichia coli isolated from urine cultures, Faculty of Medicine, Universidad Nacional Mayor de San Marcos, presented in 2018 in Lima.
ABSTRACT
Descriptive study conducted in order to determine the presence of the fimH and afa genes in urinary isolates of extended-spectrum beta-lactamases (ESBL) producing Escherichia coli. Isolates from project TO-06/09 of the Instituto Nacional de Salud del Niño in Lima, Peru were used. A total of 75 urinary isolates of Escherichia coli were included. Gene identification was performed by polymerase chain reaction. From the 75 isolates, 74 (98.7%) were positive for the fimH gene and 6 (8.0%) were positive for the afa gene. Virulence factors produced by the fimH and afa genes were evident in urinary isolates of ESBL producing Escherichia coli.
Keywords: Uropathogenic Escherichia coli; Virulence Factors; beta-Lactamases; Peru (source: MeSH NLM).
INTRODUCTION
Urinary
tract infections (UTIs) in children can affect both the upper and lower urinary
tract, causing several urinary disorders, such as cystitis and pyelonephritis.
During childhood, approximately 6% to 8% of pediatric patients with urinary
symptoms have a UTI (1,2). Frequency varies
according to several factors, such as age and gender. It is more common in
girls and uncircumcised boys. UTIs in children with urinary tract abnormalities
such as neurogenic bladder or vesicoureteral reflux may result in irreversible
kidney damage (3,4).
UTIs are caused by a group of microorganisms known as uropathogens, which can minimize the host’s immune response
and invade the urinary system with uropathogenic Escherichia
coli (UPEC), causing 85% of episodes of acute cystitis in humans. This pathotype has virulence factors that allow it to adhere to
and invade tissues, besides, these factors determine the capacity for
infection, chronicity, recurrence and the possibility of dissemination to other
tissues (5,6).
Among the most frequent virulence factors of UPEC are fimbriae (P and
type 1); adhesins, such as fimH,
S, M FIC, Dr/afa, Sfa; and systems for the uptake of iron (aerobactins), alpha‑hemolysin and
other enzymes with protease activity (5). fimH adhesin is
present in more than 80% of Escherichia coli strains that cause UTIs.
This adhesin is responsible for generating the
adhesion of the bacteria to the urinary tissue, thus favoring colonization and
subsequent invasion of the urothelium (6).
afa adhesin appears in no more than 40% of UPEC but is a key
element in the development of infections in children and pregnant women because
of its ability to cause complications (7). In addition,
antimicrobial susceptibility profiles of UPECs need to be constantly updated to
provide adequate empirical treatment for urinary tract infections, as these may
vary according to origin, geographical region or institution (8).
The main mechanism of resistance to beta-lactams in enterobacteria is the production of extended-spectrum beta-lactamase (ESBL). Since ESBL have the capacity to hydrolyze most of the beta-lactams (except carbapenemics and cephamycins), it is a pattern of multi-resistance, which causes a serious therapeutic problem. This explains its association with higher mortality, hospital stay and increased economic cost (9). Therefore, the objective of this study was to determine the presence of the fimH and afa genes in urinary isolates of ESBL-producing Escherichia coli.
|
KEY MESSAGES |
|
Motivation for the study: Adhesins (such as fimH and afa) are responsible for the colonization, invasion and chronicity of uropathogenic Escherichia coli infections. Main findings: In urinary isolates of ESBL‑producing Escherichia coli, the fimH gene was present in 98.7%, and the afa gene, in 8.0% of the samples. Implications: The presence of fimH and afa in isolates of ESBL-producing Escherichia coli from pediatric patients could indicate a relationship between adhesins and age group. A possible relationship was found between being not sensitive to amikacin and the afa gene. |
THE STUDY
A
descriptive study conducted to evaluate urinary isolates of ESBL-producing Escherichia
coli (the bacteria were collected between August 2012 and January 2013)
from the strain obtained from project TO-06/09 of the Instituto
Nacional de Salud del Niño (Molecular detection and
characterization of extended-spectrum beta lactamases in E. coli and K.
pneumoniae isolated at the Instituto Nacional de Salud del Niño). A total of 75 consecutive non-repeated
isolates were recovered from urine samples from pediatric patients in the
inpatient and outpatient departments.
Molecular detection was performed at the Laboratory of Molecular Epidemiology and Genetics of the Instituto de Medicina Tropical Daniel A. Carrión - Universidad Nacional Mayor de San Marcos (UNMSM). Total DNA was used as a mold. The fimH gene was amplified by the polymerase chain reaction (PCR) method according to Tolentino’s protocol (10). For the afa gene, a protocol was standardized in this study considering the concentration of primers, Taq polymerase DNA, hybridization temperature and mold DNA concentration.
IBM SPSS Statistics for Windows, version 25 (IBM Corp., Armonk, NY, USA) was used to report absolute and relative frequencies for the variables of interest that were obtained from the strain database.
The
study protocol was approved by the UNMSM School of Medical Technology. The
study follows the good practice and ethics in biomedical research guidelines.
RESULTS
Out
of the 75 ESBL-producing Escherichia coli isolates from urine cultures,
74 (98.7%) were positive for the fimH gene,
and 6 (8.0%) were positive for the afa gene.
In the descriptive analysis of the variables (Table
1), the frequency of fimH gene-producing Escherichia coli
according to its origin was 31.1% in a hospital setting and 68.9% in the
community. For the afa gene, the frequency was
16.7% in a hospital setting and 83.3% in the community. Out of the total number
of isolations, 54 (72%) were female and the median age was 3 years. No link was
found between the virulence genes and the gender, age and location variables.
The antibiotic susceptibility profile from ESBL-producing Escherichia coli
isolates is shown in
Figure 2. The relationship between the presence of
virulence genes and non-sensitivity to antibiotics was evaluated. An
association was found between non-sensitivity to amikacin and the presence of
the afa gene (Table
2).
Table 1.
General distribution of Escherichia coli virulence genes.
|
Characteristics |
afa gene |
fimH gene |
||
|
Yes (%) |
No (%) |
Yes (%) |
No (%) |
|
|
Gender |
|
|
|
|
|
Male |
2 (9.5) |
90 (90.5) |
21 (100) |
- |
|
Female |
4 (7.4) |
50 (92.6) |
53 (98.2) |
1 (1.8) |
|
Age (years) |
- |
|
|
|
|
<1 |
1 (5.0) |
19 (95.0) |
20 (100) |
- |
|
1 |
1 (7.7) |
12 (92.3) |
13 (100) |
- |
|
2 |
- |
3 (100) |
3 (100) |
- |
|
3 |
1 (16.7) |
5 (83.3) |
5 (83.3) |
1 (16.7) |
|
4 |
1 (20.0) |
4 (80.0) |
5 (100) |
- |
|
6 |
- |
6 (100) |
6 (100) |
- |
|
7 |
- |
7 (100) |
7 (100) |
- |
|
8 |
2 (40.0) |
3 (60.0) |
5 (100) |
- |
|
9 |
- |
1 (100) |
1 (100) |
- |
|
11 |
- |
1 (100) |
1 (100) |
- |
|
12 |
- |
2 (100) |
2 (100) |
- |
|
13 |
- |
5 (100) |
5 (100) |
- |
|
14 |
- |
1 (100) |
1 (100) |
- |
|
Localization |
|
|
|
|
|
Community |
|
|
|
|
|
Outpatient |
4 (9.1) |
40 (90.9) |
44 (100) |
- |
|
Emergency |
1 (12.5) |
7 (87.5) |
7 (87.5) |
1 (12.5) |
|
Hospital |
|
|
|
|
|
Internal medicine |
- |
5 (100) |
5 (100) |
- |
|
Neonatology ICU |
- |
4 (100) |
4 (100) |
- |
|
Orthopedics |
- |
2 (100) |
2 (100) |
- |
|
Pneumology |
- |
1 (100) |
1 (100) |
- |
|
ICU |
- |
2 (100) |
2 (100) |
- |
|
Neurology |
- |
2 (100) |
2 (100) |
- |
|
Nephrology |
- |
2 (100) |
2 (100) |
- |
|
Cardiology |
1 (50.0) |
1 (50.0) |
2 (100) |
- |
|
Surgery |
- |
1 (100) |
1 (100) |
- |
|
Urology |
- |
1 (100) |
1 (100) |
- |
|
Gynecology |
- |
1 (100) |
1 (100) |
- |
ICU: Intensive Care Unit
Table 2. Virulence genes according to antibiotic susceptibility
|
Antibiotics |
afa gene |
fimH gene |
||
|
Yes (%) |
No (%) |
Yes (%) |
No (%) |
|
|
Amikacin |
|
|
|
|
|
Not sensible |
3 (33.3) |
6 (66.7) * |
9 (100) |
- |
|
Sensible |
3 (4.5) |
63 (95.5) |
65 (98.5) |
1 (1.5) |
|
Gentamicin |
|
|
|
|
|
Not sensible |
3 (6.7) |
42 (93.3) |
45 (100) |
- |
|
Sensible |
3 (10.0) |
27 (90.0) |
29 (96.7) |
1 (3.3) |
|
Ciprofloxacin |
|
|
|
|
|
Not sensible |
6 (8.8) |
62 (91.2) |
67 (98.5) |
1 (1.5) |
|
Sensible |
- |
7 (100) |
7 (100) |
|
|
Imipenem |
|
|
|
|
|
Not sensible |
- |
- |
- |
- |
|
Sensible |
6 (8.0) |
69 (92.0) |
74 (98.7) |
1 (1.3) |
|
Meropenem |
|
|
|
|
|
Not sensible |
- |
- |
- |
- |
|
Sensible |
6 (8.0) |
69 (92.0) |
74 (98.7) |
1 (1.3) |
|
Trimethoprim-sulfamethoxazole |
|
|
|
|
|
Not sensible |
5 (9.3) |
49 (90.7) |
53 (98.5) |
- |
|
Sensible |
1 (4.8) |
20 (95.2) |
21 (100) |
- |
|
Nitrofurantoin |
|
|
|
|
|
Not sensible |
1 (14.3) |
6 (85.7) |
7 (100) |
- |
|
Sensible |
5 (7.4) |
63 (92.6) |
67 (98.5) |
1 (1.5) |
*p-value < 0.05 with Chi-square test
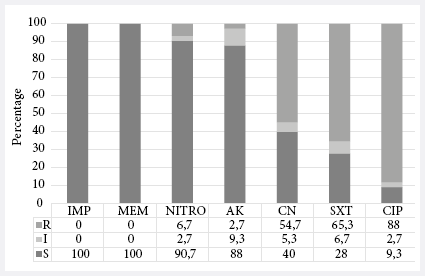
Figure 2.
Antimicrobial Resistance Profile of ESBL-producing Escherichia coli
isolates (n=75). R: resistant; I: intermediate; S: susceptible; IMP: imipenem;
MEM: meropenem; NITRO: nitrofurantoin; AK: amikacin;
CN: gentamicin; SXT: sulfamethoxazole-trimethoprim; CIP: ciprofloxacin.
In the PCR standardization for the afa
gene, temperature gradient was performed and the best hybridization temperature
was obtained at 62 °C, with a minimum concentration of primers
at 1 μM, and of Taq polymerase DNA at 0.5 U/rx. The minimum concentration
of DNA detectable by the test was 400 pg/dL (Figure 1).
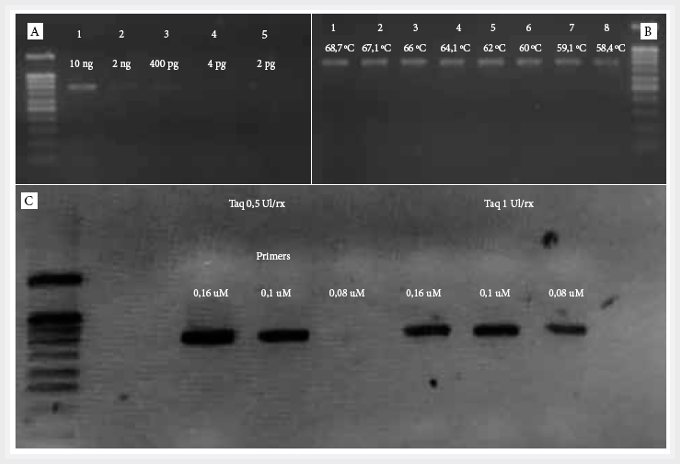
Figure 1.
Standardization of the DNA polymerase chain reaction for the afa gene in a 2% ladder agarose gel of 100 bp. A. Concentration gradient to evaluate the PCR
sensitivity for the afa gene (molecular weight
of the afa gene 750 bp).
The minimum concentration detected was 400 pg. B. Temperature gradient for the
standardization of the hybridization temperature. C.
Simultaneous standardization of the concentration of primers and Taq polymerase DNA.
It was concluded that the optimal concentration of primers is 0.1 μM with 0.5
IU Taq concentration.
DISCUSSION
The
results of this study show that 98.7% of ESBL‑producing Escherichia coli
isolates recovered from a pediatric population presented the fimH gene. This finding is similar to that reported
by Kim, et al. (11), who observed the presence of fimH adhesin in the total
number of Escherichia coli isolates from pediatric urine cultures and
found a relationship between this adhesin and the phylogroups B2 and D. Likewise, Tabasi,
et al. (12) found, after studying isolates from adult
patients with UTIs, that 100% of Escherichia coli isolates carried the fimH gene from isolates of UTI patients in the adult
population. Similarly, Rahdar, et al. (13)
detected the fimH gene in 95% of UPEC isolates
and found no relationship between the presence of the fimH
gene and Escherichia coli phylogroups. In
2009, Berry, et al. described the mechanism of action for fimH adhesin, which acts
and interacts with urothelium, allowing UPEC to enter
and form intracellular bacterial colonies (IBC) after the first 6 hours of
infection (14). IBCs are responsible for the recurrence, chronicity,
and formation of bacterial reservoirs in the urothelium
(15).
Regarding the afa gene, we reported a
frequency of 8.0% in the studied UPEC; this is consistent with the findings of
Ramirez (16), who reported a frequency of 8.2% of the afa gene in multiresistant
UPEC strains. In contrast, Tabasi, et al. (12),
reported a frequency of 29.5% for the afa
gene, and a relationship between cystitis and recurrent infections with the
presence of the afa gene. On the other hand, Servin considers that the presence of the afa gene (subtype afaE)
is more frequent in Escherichia coli that causes
pyelonephritis (7). Tajbakhsh, et al.
reported that 32% of UPEC isolates had the afa
gene, and found a significant association between the presence of afa and biofilm production (p<0.05), a
characteristic that was also associated with the presence of beta-lactamases (17).
In 2017 Souza, et al. reported a 9% frequency of the afa gene in UPEC isolates (18) but found
no significant association between the gene and the phylogroup,
nor with the gender of the patient from which it was
isolated. Furthermore, they showed that the presence of the afa
gene and being not sensitive to amikacin were significantly associated. Which differs from what was reported by Malekzadegan,
et al. (19), who found no association between the afa gene and no being sensitive to amikacin, but did
find an association between the presence of the afa
gene and the production of ESBL.
The study has some limitations that are relevant to address. The results
obtained correspond to a single center, which could differ according to each
population and institution. Furthermore, no other virulence factors of
importance in the pediatric population were sought, nor their possible
relationship with resistance markers.
In conclusion, the presence of virulence factors produced by the genes fimH and afa was evidenced in urinary isolates of ESBL-producing Escherichia coli. Besides, the standardization and optimization of PCR for the detection of the afa gene performed satisfactorily.
Acknowledgements: To the staff of the Laboratory of Molecular Epidemiology and Genetics of the Universidad Nacional Mayor de San Marcos for their continued support.
REFERENCES
1. Kaufman J, Temple-Smith M, Sanci L. Urinary tract infections in children: an overview of diagnosis and management. BMJ Paediatr Open. 2019;3(1):e000487. doi: 10.1136/bmjpo-2019-000487.
2. Leung AKC, Wong AHC, Leung AAM, Hon KL. Urinary Tract Infection in Children. Recent Pat Inflamm Allergy Drug Discov. 2019;13(1):2-18. doi: 10.2174/1872213X13666181228154940.
3. Tullus K. Fifteen-minute consultation: ¿Why and how do children get urinary tract infections?. Arch Dis Child Educ Pract Ed. 2019;104(5):244-7. doi: 10.1136/archdischild-2018-315023.
4. Asociación Española de Pediatría. Protocolos diagnósticos y terapéuticos en Pediatría: Infectología pediátrica [Internet]. España: ERGON; 2011[citado el 13 de marzo de 2020]. Disponible en: https://www.aeped.es/documentos/protocolos-infectologia-en-revision.
5. Terlizzi ME, Gribaudo G, Maffei ME. UroPathogenic Escherichia coli (UPEC) Infections: Virulence Factors, Bladder Responses, Antibiotic, and Non-antibiotic Antimicrobial Strategies. Front Microbiol. 2017;8:1566. doi: 10.3389/fmicb.2017.01566.
6. Najafi A, Hasanpour M, Askary A, Aziemzadeh M, Hashemi N. Distribution of pathogenicity island markers and virulence factors in new phylogenetic groups of 57 uropathogenic Escherichia coli isolates. Folia Microbiol (Praha). 2018;63(3):335- 43. doi: 10.1007/s12223-017-0570-3.
7. Servin AL. Pathogenesis of human diffusely adhering Escherichia coli expressing Afa/Dr adhesins (Afa/Dr DAEC): current insights and future challenges. Clin Microbiol Rev. 2014;27(4):823-69. doi: 10.1128/CMR.00036-14.
8. Warren J, Abrutyn E, Hebel J, Johnson J, Schaeffer A, Stamm W. Guidelines for antimicrobial treatment of uncomplicated acute bacterial cystitis and acute pyelonephritis in women. Infectious Diseases Society of America (IDSA). Clin Infect Dis. 1999;29(4):745-58. doi: 10.1086/520427.
9. Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol. 2005;18(4):657-86. doi: 10.1128/CMR.18.4.657-686.2005.
10. Tolentino E. Detección genotípica de los factores de virulencia: Fimbria tipo 1, Fimbria P y alfa hemolisina en Escherichia coli aisladas de urocultivos [tesis de pregrado]. Lima: Facultad de Medicina, Universidad Nacional Mayor de San Marcos; 2015. Disponible en: http://cybertesis.unmsm.edu.pe/bitstream/handle/cybertesis/7519/Tolentino_le%20-%20Resumen.pdf?sequence=1&isAllowed=y.
11. Kim D, Subhadra B, Kang H, Woo K, Kim J, Son Y, et al. Virulence properties of uropathogenic Escherichia coli isolated from children with urinary tract infection in Korea. Genes Genomics. 2018;40(6):625-34. doi: 10.1007/s13258-018-0664-6.
12. Tabasi M. Genotypic Characterization of Virulence Factors in Escherichia coli Isolated from Patients with Acute Cystitis, Pyelonephritis and Asymptomatic Bacteriuria. J Clin Diagn Res. 2016;10(2):1-7. doi: 10.7860/JCDR/2016/21379.9009.
13. Rahdar M, Rashki A, Miri HR, Rashki Ghalehnoo M. Detection of pap, sfa, afa, foc, and fim Adhesin-Encoding Operons in Uropathogenic Escherichia coli Isolates Collected From Patients With Urinary Tract Infection. Jundishapur J Microbiol. 2015;8(8):e22647. doi: 10.5812/jjm.22647.
14. Berry R, Klummp D, Schaeffer A. Urothelial cultures support Intracellular Bacterial Community Formation by uropathogenic Escherichia coli. Journal Infect Inmun. 2009; 77(7):2762-72. doi: 10.1128/IAI.00323-09.
15. Hanna T, Totsika M, Mansfield K. Host-Pathogen Checkpoints and Population Bottlenecks in Persistent and Intracellular Uropathogenic E. coli Bladder Infection. FEMS Microbiol Rev. 2012;36(3):616-648. doi: 10.1111/j.1574-6976.2012.00339.x.
16. Ramírez-Castillo FY, Moreno-Flores AC, Avelar-González FJ, Márquez-Díaz F, Harel J, Guerrero-Barrera AL. An evaluation of multidrug-resistant Escherichia coli isolates in urinary tract infections from Aguascalientes, Mexico: cross-sectional study. Ann Clin Microbiol Antimicrob. 2018;17(1):34. doi: 10.1186/s12941-018-0286-5.
17. Tajbakhsh E, Ahmadi P, Abedpour-Dehkordi E, Arbab-Soleimani N, Khamesipour F. Biofilm formation, antimicrobial susceptibility, serogroups and virulence genes of uropathogenic E. coli isolated from clinical samples in Iran. Antimicrob Resist Infect Control. 2016;5(11):1-8. doi: 10.1186/s13756-016-0109-4.
18. de Souza da-Silva AP, de Sousa VS, Martins N, da Silva RC, Bonelli RR, Riley LW, et al. Escherichia coli sequence type 73 as a cause of community acquired urinary tract infection in men and women in Rio de Janeiro, Brazil. Diagn Microbiol Infect Dis. 2017;88(1):69-74. doi: 10.1016/j.diagmicrobio.2017.01.024.
19. Malekzadegan Y, Khashei R, Sedigh Ebrahim-Saraie H, Jahanabadi Z. Distribution of virulence genes and their association with antimicrobial resistance among uropathogenic Escherichia coli isolates from Iranian patients. BMC Infect Dis. 2018;18(1):572. doi: 10.1186/s12879-018-3467-0.
Citation: Matta-Chuquisapon J, Valencia-Bazalar E, Marocho-Chahuayo L, Gonzales-Escalante E, Sevilla-Andrade CR. Presence of fimH and afa genes in urinary isolates of extended-spectrum betalactamases producing Escherichia coli in Lima, Peru. Rev Peru Med Exp Salud Publica. 2020;37(2):282-6. doi: https://doi.org/10.17843/rpmesp.2020.372.4829.
Correspondence to: Jose Fernando Matta Chuquisapon; Calle José Santos Chocano 199, Ciudad Universitaria, Bellavista, Callao; josematta5511295@gmail.com.
Authorship contributions:
All authors have
participated in the research idea, conception of the article, data collection
and study material, and writing of the article. All authors approved the final
version of the manuscript and assume responsibility for its content.
Conflicts of Interest: The authors have no
conflict of interest to declare.
Funding sources: Self-funded.
Received: 23/09/2019
Approved: 15/04/2020
Online: 10/06/2020